Directed Graphs: Setup and Representations¶
Denote \(G = (V, E)\) a graph, where \(V\) are the vertices or nodes and \(E\) are the links or edges.
This notebook shows how to setup directed graphs, how to access different graph respresentations
and how to specify labels to indices mappings.
A directed graph is characterized by the direction of the edge flow, i.e., units can only travel along the edge in one direction.
Consider the following directed graph:
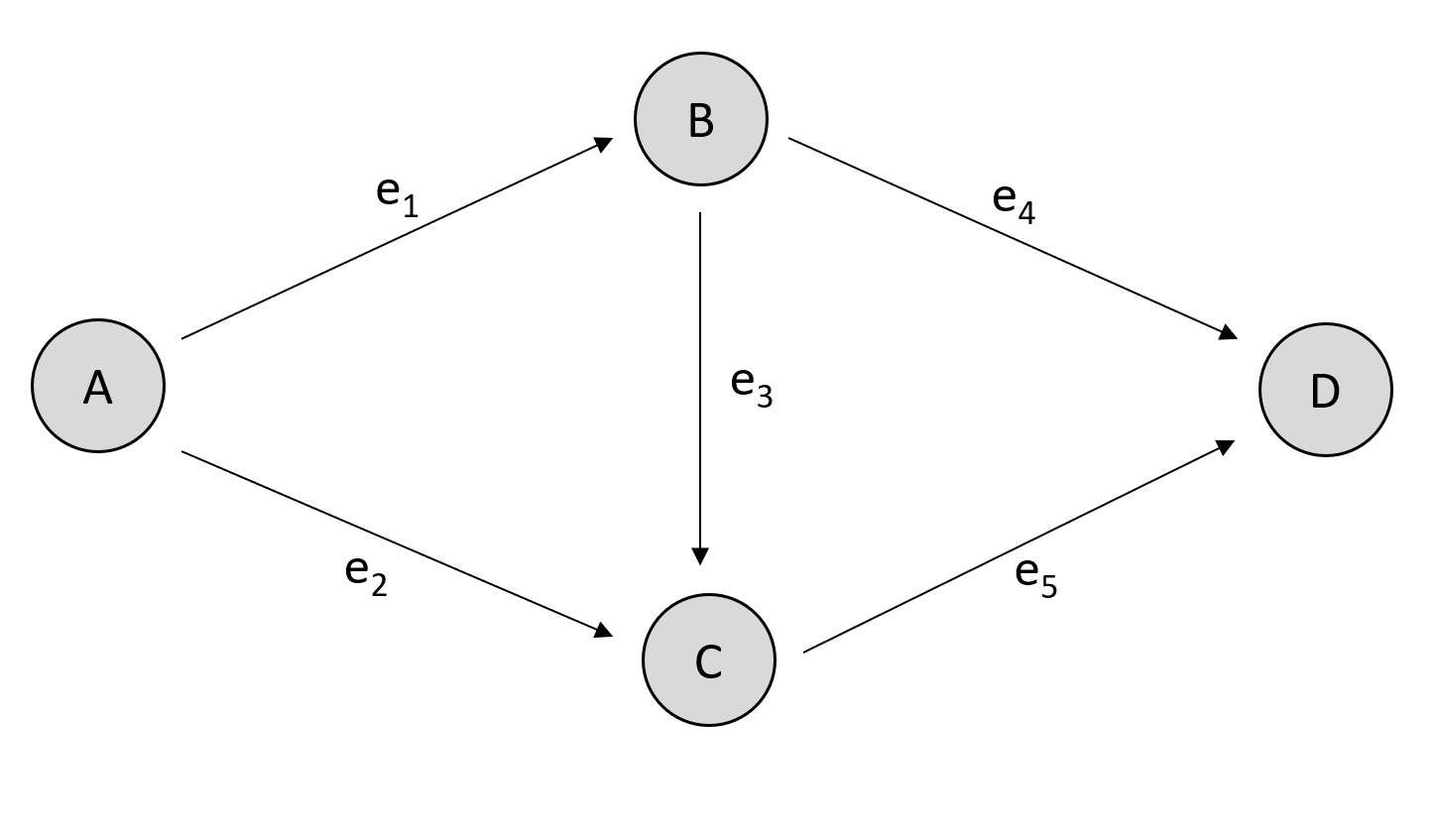
Simple directed graph.¶
that connectects the four vertices \(A, B, C, D\) with the five directed links \(e_1, e_2, e_3, e_4\) and \(e_5\).
1 Setup¶
We can setup the above graph with paminco:
import numpy as np
import paminco
edge_data = np.array([
["A", "B"],
["A", "C"],
["B", "C"],
["B", "D"],
["C", "D"],
])
graph = paminco.Network(edge_data)
graph.edges.to_df()
| source_lbl | target_lbl | s | t | lb | ub | |
|---|---|---|---|---|---|---|
| 0 | A | B | 0 | 1 | 0.0 | inf |
| 1 | A | C | 0 | 2 | 0.0 | inf |
| 2 | B | C | 1 | 2 | 0.0 | inf |
| 3 | B | D | 1 | 3 | 0.0 | inf |
| 4 | C | D | 2 | 3 | 0.0 | inf |
2 Representations¶
A graph can be represented by its \(n \times n\) adjacency matrix, which store as a sparse matrix:
graph.adjacency_matrix().toarray()
array([[0., 1., 1., 0.],
[0., 0., 1., 1.],
[0., 0., 0., 1.],
[0., 0., 0., 0.]])
or its \(n \times m\) incidence matrix
graph.incidence_matrix().toarray()
array([[-1, -1, 0, 0, 0],
[ 1, 0, -1, -1, 0],
[ 0, 1, 1, 0, -1],
[ 0, 0, 0, 1, 1]], dtype=int32)
3 Edge bounds¶
Depending on the specific problem, each edge may only allow a certain maximum of flow on the edge, i.e., it has an upper bound. By default these bounds are \((0, \infty)\) for directed edges. However, it is possible to pass individual flow bounds for each edge:
edge_labels = np.array([
["A", "B"],
["A", "C"],
["B", "C"],
["B", "D"],
["C", "D"],
])
edge_bounds = np.array([
[1, 100],
[4, 5],
[300, 400],
[0, 80000],
[12, 12.1],
])
edge_data = (edge_labels, edge_bounds)
graph = paminco.Network(edge_data)
graph.edges.to_df()
| source_lbl | target_lbl | s | t | lb | ub | |
|---|---|---|---|---|---|---|
| 0 | A | B | 0 | 1 | 1.0 | 100.0 |
| 1 | A | C | 0 | 2 | 4.0 | 5.0 |
| 2 | B | C | 1 | 2 | 300.0 | 400.0 |
| 3 | B | D | 1 | 3 | 0.0 | 80000.0 |
| 4 | C | D | 2 | 3 | 12.0 | 12.1 |
4 Labels to indices mapping¶
In a network, the nodes objects handles the mapping of labels to indices, which influences the graph representation.
The mapping is accessed by:
graph.nodes.lbl2id
{'A': 0, 'B': 1, 'C': 2, 'D': 3}
It is possible to set a mapping in two ways:
pass in indices in node data
specfiy mapping for edge constructor
4.1 Pass indices in node data¶
labels = ["A", "B", "C", "D"]
indices = [3, 1, 0, 2]
zone = False
xy = [(0, 0), (4, 3), (8, 8), (1, 9)]
node_data = (labels, indices, xy, zone)
g = paminco.Network(edge_data, node_data)
g.nodes.lbl2id
{'C': 0, 'B': 1, 'D': 2, 'A': 3}
E.g., compare the indicence matrices, where the specified mapping changes to row order:
g.incidence_matrix().toarray()
array([[ 0, 1, 1, 0, -1],
[ 1, 0, -1, -1, 0],
[ 0, 0, 0, 1, 1],
[-1, -1, 0, 0, 0]], dtype=int32)
graph.incidence_matrix().toarray()
array([[-1, -1, 0, 0, 0],
[ 1, 0, -1, -1, 0],
[ 0, 1, 1, 0, -1],
[ 0, 0, 0, 1, 1]], dtype=int32)
For more information see documentation of Nodes:
print(paminco.net.Nodes.__doc__[:1010]) # smaller version of paminco.net.Nodes?
Class that contains the nodes/vertices of a network.
A Nodes object can be instantiated in several ways:
Nodes(n)
where ``n`` is a Nodes object.
Nodes(nodes)
where ``nodes`` is array_like and contains either node
labels or node indices. If no indices are given, they
are set automatically.
Nodes(nodes, zone)
where ``nodes`` and ``zone`` are array_like of shape (n, ).
``zone`` must be boolean array denoting if a node is a
zone, mostly used for traffic networks.
Nodes(nodes, xy, zone)
where ``nodes`` and ``zone`` are array_like of shape (n, )
and ``xy`` is array_like of shape (n, 2) and contains the
coordinates of the nodes.
Nodes(node_labels, node_indices, xy, zone)
where ``node_labels`` and ``node_indices`` and ``zone``
are are array_like of shape (n, ) and ``xy`` is array_like
of shape (n, 2).
4.2 Specifing label mapping in edges¶
We can achieve the same by specfying a map_labels_to_indices mapping in edges:
edge_data = np.array([
["A", "B"],
["A", "C"],
["B", "C"],
["B", "D"],
["C", "D"],
])
mapping = {'C': 0, 'B': 1, 'D': 2, 'A': 3}
g2 = paminco.Network(edge_data, kw_edge={"map_labels_to_indices": mapping}, dtype_int=np.int64)
g2.incidence_matrix().toarray() - g.incidence_matrix().toarray()
array([[0, 0, 0, 0, 0],
[0, 0, 0, 0, 0],
[0, 0, 0, 0, 0],
[0, 0, 0, 0, 0]], dtype=int32)